
Google DeepMind’s Latest AI Model Is Poised to Revolutionize Drug Discovery
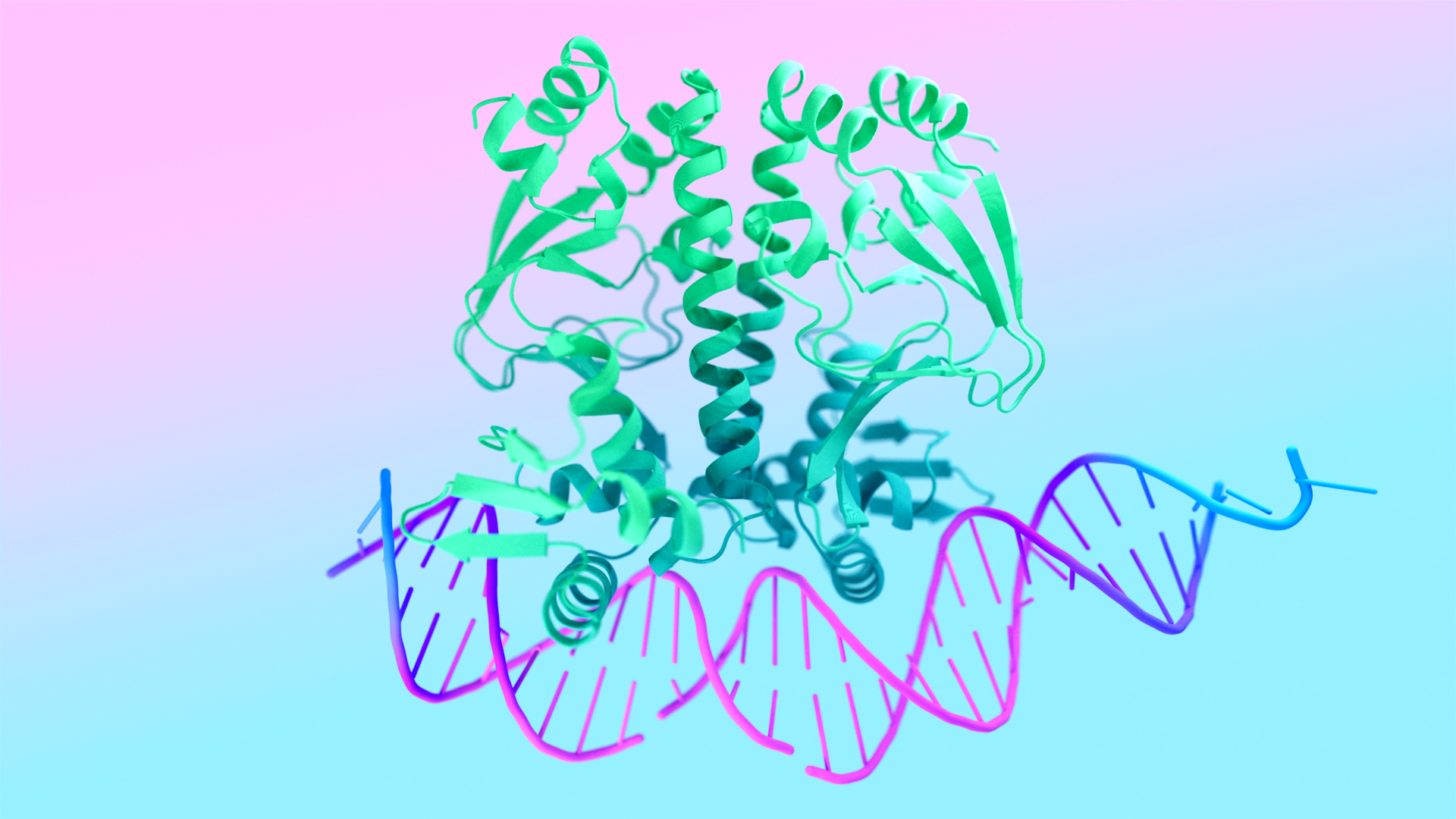
Researchers at Google DeepMind have developed AlphaFold 3, an AI model that can predict the structure of and interactions between biological molecules including proteins, DNA and RNA, and small molecules that could function as drugs. Google DeepMind will make the model available for non-commercial use through AlphaFold server. The landmark innovation, the details of which were published in the journal Nature on May 8, is likely to dramatically accelerate biological research.
“It’s a big milestone for us today, announcing AlphaFold 3,” said Demis Hassabis, CEO of Google DeepMind, at a briefing on May 7 announcing the breakthrough. “Biology is a dynamic system and you have to understand how properties of biology emerge through the interactions between different molecules in the cell. You can think of AlphaFold 3 as our first big step towards that.”
[time-brightcove not-tgx=”true”]
The AI system is a descendant of previous AlphaFold models built by Google DeepMind that essentially solved the problem of predicting the three-dimensional structure of a protein from its amino acid structure. Google DeepMind’s first AlphaFold model, announced in 2018, attempted to predict protein structures, coming first in an international protein structure prediction competition. AlphaFold 2, released in 2020, significantly improved on the first’s protein structure accuracy predictions.
AlphaFold 3 goes further by predicting the structures of almost all biological molecules and modeling the interactions between those molecules. While researchers have long developed specialized computational methods for modeling interactions between specific types of biological molecules, AlphaFold 3 marks the first time that a single system has been able to predict the interactions between nearly all molecular types with state-of-the-art performance.
The properties and functions of molecules in biological systems are typically a result of how they interact with other molecules. Using experiments to understand molecular interactions can take years of research time and be prohibitively expensive. If these interactions can instead be estimated computationally with sufficient accuracy, then biological research can be dramatically accelerated. For example, if researchers believe that a molecule that binds to a specific site on a certain protein would be a promising drug candidate, they can use computational systems such as AlphaFold 3 to test potential drug molecules.
“AlphaFold continues to get better, and increasingly more relevant for biological investigations,” said Paul Nurse, the Nobel Prize-winning geneticist and chief executive and director of the London-based biomedical research center the Francis Crick Institute, in a statement accompanying the Google DeepMind announcement. “This third version will enable increased accuracy in predicting the structures of complexes between different macromolecules, as well as associations between macromolecules, small molecules and ions.”
Google DeepMind was founded as DeepMind in 2010 by Hassabis, along with Google DeepMind Chief AGI Scientist Shane Legg and Mustafa Suleyman. (Suleyman is now CEO at Microsoft AI, Microsoft’s consumer AI products and research organization.) DeepMindwas acquired by Google in 2014, and in 2023 Google merged DeepMind with Google Brain, another Google AI division, to form Google DeepMind, putting an end to efforts by DeepMind’s leadership to secure greater autonomy from their parent company.
In addition to the AlphaFold family of AI systems, Google DeepMind has made several breakthroughs that use AI to further science and technology. In 2022 the company released an AI system that can discover novel algorithms, and in 2023 it released an AI model that could forecast the weather with unprecedented accuracy. Also in 2023, Google DeepMind released an AI model that it claims accurately predicts the structures of materials, although the utility of this model has since been called into question by independent researchers.
In 2021, Google parent company Alphabet announced the creation of Isomorphic Labs, which aims to take an AI-first approach to drug discovery. Researchers from Isomorphic Labs contributed to the development of AlphaFold 3 and, while AlphaFold Server can be used by anyone from non-commercial research, researchers at Isomorphic Labs will have exclusive access to AlphaFold 3 for commercial use.
“We’ve been using AlphaFold 3’s capabilities day-to-day in our drug design programmes,” said Max Jaderberg, chief AI officer at Isomorphic labs, at the announcement briefing. “We’re already seeing that potential to accelerate, improve, and ultimately transform the way that we do drug discovery, and it’s really because of the new level of accuracy of this model, and the increased breadth of biomolecules that this model is able to predict, that really enables that for us.”
Get the latest work and career updates delivered straight to your inbox by subscribing to our magazine category today. Stay informed and ahead of the game with Subscrb.
The content on this website has been curated from various sources and is for informational purposes only. We do not claim ownership of any of the content posted here, all rights belong to their respective authors. While we make every effort to ensure that the information is accurate and up-to-date, we cannot guarantee its completeness or accuracy. Any opinions or views expressed on this website are solely those of the original authors and do not necessarily represent our own. We do not endorse or take responsibility for the content or actions of external websites or individuals linked from this website. Any reliance on the information provided on this website is done at your own risk. Please note that this article was originally seen on the source website TIME, by the author Will Henshall
-
SALE!




Forbes Asia Magazine Subscription
From: RM220 / year -
SALE!


Fortune Magazine Subscription
From: RM118 / year -
OUT OF STOCK




The Economist Magazine Subscription
From: RM1530 / year -


Inc. Magazine Magazine Subscription
From: RM22 / year -


Consumer Reports Magazine Subscription
From: RM22 / year -


Harvard Business Review Magazine Subscription
From: RM83 / month -


Entrepreneur’s Startups Magazine Subscription
From: RM4 / year -


BILLIONAIRE Magazine Subscription
From: RM131 / year



